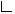 net.sourceforge.nite.datainspection.timespan.SegmentBasedInspection
net.sourceforge.nite.datainspection.timespan.SegmentBasedInspection
|
|||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | ||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | ||||||||
java.lang.Objectnet.sourceforge.nite.datainspection.timespan.SegmentBasedInspection
public class SegmentBasedInspection
Inspection tool for timeline segmentations (gapped or non gapped) that investigates whether two annotators identified same segments. Segments detected by two annotators are taken to be the same if they differ in start or end time at most a (configurable) threshold 'th'. Kappa and alpha are calculated by comparing the labeling for each aligned pair of segments.

Varying threshold 'th' gives information about the precision with which two annotators identified the same segments. Note however that 'th' should be low enough compared to the segment lengths in the annotations.
========
===Aligning f vs e
---
- th 0.0
9 aligned segments
For ann1: 2%.
For ann2: 3%.
---
- th 1.5
242 aligned segments
For ann1: 74%.
For ann2: 90%.
---
- th 3.0
248 aligned segments
For ann1: 76%.
For ann2: 92%.
The list above gives an example output obtained from the SegmentBasedInspection tool, for the FOA layer of the AMI corpus, with several variations of 'th'. Again, we look at the visualisations of the alignments, because of course we are afraid of running into the same 'th' sensitivity that we had with the boundary alignments. The following image show an alignment produced using the tool SegmentBasedInspection. Actually (but for now you'll have to take my word for it...) it turns out that this alignment is for this annotation a lot better when it operates on segments instead of boundaries (see the boundary based inspection for example alignments). Furthermore it is less sensitive to too high threshold values. In this annotation even a treshold of 3 seconds leads to almost only correct alignments (compare to the maximum adequate threshold of about 0.3 seconds for the boundary alignment).
Remember why you do reliability analysis. The reason to do reliability analysis is not only to reach a conclusion like "the alpha reliability of my annotation is 0.8 so it is good enough". The main reason to do this data inspection is to figure out the strong points and the weak points of your annotations. Hopefully identifying those strong and weak points helps you improve the annotation protocol, or decided what you can and cannot do with the data resulting from your annotation protocol.

The kind of alignment mistake described above is often unavoidable. When they don't happen too often, they are also not so much of a problem, since the data inspection packages aim at getting an idea of the quality of the corpus, not at getting an absolute truth about correctness of annotations. In some cases however it is quite easy to improve the alignments. In the example mentioned above, the hand gesture annotation of AMI, the no_comm_hand label is a kind of "background label" which is used whenever there is no communicative hand gesture. Since the alignment procedure aims at aligning the segments where two annotators agree on the occurrence of (in this case) a gesture, it seems sensible to leave out these background segments from the analysis. This is supported by the 'isIgnoreP' parameter in the constructor of the SegmentBasedInspection tool. If this parameter is non-null, all segments for which this predicate evaluates true are ignored in the analysis. If we run the alignment for the AMI hand gestures anew while defining "no_comm_hand" as an to-be-ignored label we get the alignment shown below.

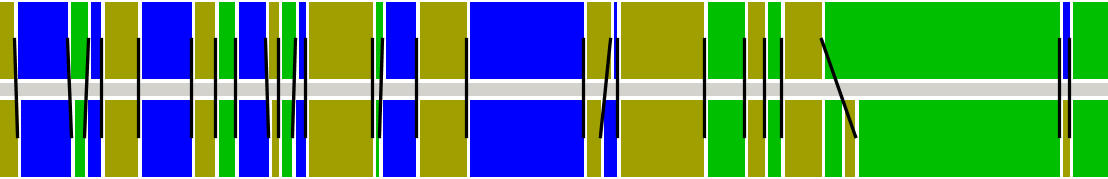
SegmentBasedInspection; some information needed. You must provide a DistanceMetric and a NOMElementToValueDelegate to the constructor of the SegmentBasedInspection tool. These are used to calculate Krippendorff Alpha on the aligned segments. These values are reported on the System.out. Furthermore confusion tables are shown on screen for every pair of annotators.
For the AMI hand gesture annotation, the output is as follows (Alpha reported for all aligned hand gestures using the string type label as Value and a BooleanMetric as distance metric):
Deriving matrices for BrigitteGreenwood vs pistek
==============================================================
Alpha = 0.7226277372262774 for threshold 0.5
==============================================================
Alpha = 0.8085808580858086 for threshold 1.5
==============================================================
Alpha = 0.8171091445427728 for threshold 2.5
Deriving matrices for BrigitteGreenwood vs xsobol03
==============================================================
Alpha = 0.7428571428571429 for threshold 0.5
==============================================================
Alpha = 0.7128712871287128 for threshold 1.5
==============================================================
Alpha = 0.7256637168141593 for threshold 2.5
| Field Summary | |
|---|---|
java.lang.String |
agentName
the name of the agent for which you want to analyse the annotations. |
java.lang.String |
codingName
the name of the Coding in which the boundaries are to be found |
java.lang.String |
corpusName
corpus |
java.lang.String |
observationName
observation |
java.lang.String |
segmentElementName
the name of the Elements in the Layer in that Coding in which the boundaries are to be found |
java.lang.String |
segmentsLayer
the name of the Layer in that Coding in which the boundaries are to be found |
| Constructor Summary | |
|---|---|
SegmentBasedInspection(java.lang.String c,
java.lang.String o,
java.lang.String codingName,
java.lang.String segmentsLayer,
java.lang.String segmentElementName,
java.lang.String agentName,
NOMElementToTextDelegate segmentToText,
Predicate isIgnoreP,
NOMElementToValueDelegate segmentToValue,
DistanceMetric labelDistanceMetric,
double thMin,
double thMax,
int thSteps)
|
|
| Method Summary | |
|---|---|
void |
collectAlignments(double thMin,
double thMax,
int thSteps)
Collect for all threshold values the alignments for all annotator pairs, using the SegmentAligner. |
void |
collectClassifications()
Collect for all threshold values the derived classifications for all alignments, using the SegmentAlignmentToClassificationFactory. |
void |
collectMatrices()
Collect for all Classification pairs the confusion and coincidence matrices used for calculation of reliability and inspection of confusions. |
void |
collectSegments()
Collect the segments for all annotators. |
void |
drawLegend()
|
void |
generateThresholdValues(double thMin,
double thMax,
int thSteps)
|
Clock |
getClock()
|
ClockFace |
getClockFace()
Returns the clockface of this class. |
NOMWriteCorpus |
getCorpus()
|
java.lang.String |
getCorpusName()
|
NiteMetaData |
getMetaData()
|
java.lang.String |
getObservationName()
|
void |
initReportPanel()
|
void |
renderRelations()
|
void |
renderSegments()
|
java.util.List |
search(java.lang.String query)
|
void |
showConfusionTables()
Show the confusion tables in internal frames on the desktop |
| Methods inherited from class java.lang.Object |
|---|
equals, getClass, hashCode, notify, notifyAll, toString, wait, wait, wait |
| Field Detail |
|---|
public java.lang.String corpusName
public java.lang.String observationName
public java.lang.String codingName
public java.lang.String segmentsLayer
public java.lang.String segmentElementName
public java.lang.String agentName
| Constructor Detail |
|---|
public SegmentBasedInspection(java.lang.String c,
java.lang.String o,
java.lang.String codingName,
java.lang.String segmentsLayer,
java.lang.String segmentElementName,
java.lang.String agentName,
NOMElementToTextDelegate segmentToText,
Predicate isIgnoreP,
NOMElementToValueDelegate segmentToValue,
DistanceMetric labelDistanceMetric,
double thMin,
double thMax,
int thSteps)
| Method Detail |
|---|
public void collectSegments()
public void collectAlignments(double thMin,
double thMax,
int thSteps)
public void collectClassifications()
public void collectMatrices()
public void initReportPanel()
public void renderSegments()
public void renderRelations()
public void drawLegend()
public void showConfusionTables()
public java.lang.String getCorpusName()
public java.lang.String getObservationName()
public NOMWriteCorpus getCorpus()
public NiteMetaData getMetaData()
public Clock getClock()
public ClockFace getClockFace()
public void generateThresholdValues(double thMin,
double thMax,
int thSteps)
public java.util.List search(java.lang.String query)
|
|||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | ||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | ||||||||